MicroRNAs (miRNAs) are endogenously expressed small ssRNA sequences of ~22 nucleotides in length that naturally direct gene silencing through components shared with the RNAi pathway. BLOCK-iT™ Pol II miR RNAi Technology uses expression vectors that allow flexible expression of miRNA-based knockdown cassettes driven by RNA Polymerase II (Pol II) promoters in mammalian cells.
To generate miR RNAi oligos for use with this system, you can use BLOCK-iT™ RNAi Designer to design two single-stranded DNA oligonucleotides: one encoding the target pre-miRNA (top-strand oligo) and the other its complement (bottom-strand oligo). You then anneal the top and bottom strands to generate a double-stranded oligonucleotide (ds oligo) suitable for cloning into BLOCK-iT™ Pol II miR RNAi Expression Vectors.
The top-strand oligo encoding the target pre-miRNA has the following features:
A 4-nucleotide, 5' overhang (TGCT) complementary to the vector (required for directional cloning)
A 5' G + a short 21-nucleotide antisense sequence (mature miRNA) derived from the target gene, followed by
A short spacer of 19 nucleotides to form a terminal loop and
A short sense target sequence with 2 nucleotides removed to create an internal loop
The structural features are depicted in the figure below.
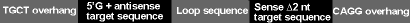
See the BLOCK-iT™ Pol II miR RNAi Expression Vector Kits manual for more information, including details about the oligo design rules used by the RNAi Designer.
To begin designing miR RNAi oligos, select the miR RNAi target design option at the top of the Designer page. Then enter a sequence for the gene that you want to target or a database accession number for that sequence, select a database to BLAST search to identify unique regions of the sequence, and specify a GC percentage range for the oligos.
You can copy and paste a nucleotide target sequence directly into the Nucleotide Sequence field, or you can enter a database accession number for the sequence in the Accession Number field. If you enter an accession number, the Designer will link to the Entrez search engine and search the crosslinked databases at the National Center for Biotechnology Information (NCBI), including GenBank and OMIM. Then the sequence associated with the accession number will be automatically downloaded into the Designer.
Note: If you enter an accession number in RefSeq format (e.g., NT_123456), the Designer will check the sequence for known single-nucleotide polymorphisms (SNPs). Designs that include SNPs will be rejected by the Designer. SNP identification will not be performed if you paste in nucleotide sequences or enter GenBank-format accession numbers.
Nucleotide Sequence: Type or paste your single-stranded target sequence into this box. The designer accepts sequences in text, FASTA, or GenBank format.
The sequence should be oriented from 5' to 3'. Because miR RNAi oligonucleotides must be highly specific, the input sequence should contain only the following 1-letter base code abbreviations: A, C, G, T, U (case insensitive). Any regions of the sequence containing ambiguous bases (e.g., N) or other letters will be skipped when generating the designs. If your sequence includes too many ambiguous bases or other letters, you will be prompted to re-enter the sequence. White spaces and numerals in the sequence will be ignored.
Note: If you are having difficulty generating designs, check your input sequence to make sure that it contains only the letters A, C, G, T, or U.
Accession Number: Enter the database accession number for your sequence in this field. The Designer will link to the Entrez search engine and search the crosslinked databases at NCBI, including GenBank and OMIM. The sequence associated with the accession number will be automatically downloaded into the Designer.
RefSeq-format accession numbers begin with two letters followed by an underscore and six digits (e.g., NT_123456). GenBank accession numbers are usually a combination of a letters and numbers, such as a single letter followed by five digits (e.g., U12345) or two letters followed by six digits (e.g., AF123456). If you enter an accession number in RefSeq format (e.g., NT_123456), the Designer will check the sequence for known single-nucleotide polymorphisms (SNPs). Designs that include SNPs will be rejected by the Designer. SNP identification will not be performed if you paste in nucleotide sequences or enter GenBank-format accession numbers.
If you entered an accession number in Step 1, in this step you specify the regions of the sequence for which you want to design your miR RNAi oligo. The default selection of Open Reading Frame (ORF) is appropriate in most cases. If you have trouble generating designs for the ORF alone, select the 5' UTR and/or 3' UTR checkboxes and redesign the oligonucleotides.
Step 3. Choose Database for BLAST
In Step 3, select a species-specific database from the pulldown list that you want to perform a BLAST (Basic Local Alignment Search Tool) search on to identify unique regions in your target sequence. Areas of homology identified by the BLAST search will be "masked" in the sequence to ensure that the target designs are highly specific.
Each BLAST search is performed against a nonredundant version of the UniGene database. The database contains representative gene sequences for the selected species.
Note: The databases used in this search contain one representation of each gene and do not include splice variants.
The Minimum GC Percentage and Maximum GC Percentage fields contain default values for the minimum and maximum percentage GC content of your miR RNAi oligos. For most sequences, this range is wide enough to generate several designs.
You can change this range if you are having difficulty generating designs, or if you know that the GC content of your oligonucleotides will fall outside this range based on the sequence. Designs outside the selected range will be rejected by the Designer.
When you have made your selections above, click on the RNAi Design button.
For information about design results, see miR RNAi Design Results.